RESEARCH ARTICLE
Pharmacogenetics — Getting Closer
Ian M. MacDonald*
Article Information
Identifiers and Pagination:
Year: 2009Volume: 3
First Page: 46
Last Page: 49
Publisher ID: TOOPHTJ-3-46
DOI: 10.2174/1874364100903010046
Article History:
Received Date: 29/1/2009Revision Received Date: 12/3/2009
Acceptance Date: 29/3/2009
Electronic publication date: 17/9/2009
Collection year: 2009

open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
Abstract
This review is written for the generalist to provide an understanding of the application of genetics to the care of patients with glaucoma and the broader concepts of personalized medicine. More specifically, the review will link advances in the genetics of glaucoma with the concepts of pharmacogenetics and its potential to improve patient care.
PHARMACOGENETICS
Genetic variation contributes to an individual’s response to human disease. This variation also underlies the response to therapy, including possible adverse effects. These are the central tenets of pharmacogenetics. Some health care leaders view pharmacogenetics as providing the potential to create personalized prescriptions; with the opportunity to improve patient compliance, reduce adverse events, and reduce the cost of managing chronic disease [1]. As a chronic disease, glaucoma seems an ideal target to apply the concepts of pharmacogenetics: creating a personalized prescription of treatment which encourages compliance, minimizes adverse events and is cost-effective.
Kalow and Gunn [2] recognized that some patients, when administered succinyl choline prior to electroconvulsant therapy, had longer lasting effects and apnea tied to their level of cholinesterase. This observation was perhaps a starting point for the discipline of pharmacogenetics and exemplifies the fact that some patients may experience a variable or irregular response to a regular treatment. These irregular responses are due to genetic differences in between patients. In this example, the irregular response to succinyl choline was determined at a genetic level; more specifically, the genetically determined pseudocholine esterase deficiency. Personalized medicine based on genetic principles, may not only avoid failures of therapy but also help prevent adverse drug reactions. In an another example, we might consider pre-treatment testing for glucose-6-phosphate dehydrogenase (G6PD) deficiency in individuals for whom primaquine or chloroquine malaria prophylaxis will be administered – trying to avoid the potential adverse side effects of the drug in a genetically at-risk population. Individuals with G6PD deficiency are at risk of developing an acute hemolytic anemia when these drugs are given.
In an editorial by the leadership at Duke University [3], these words were used: “Information systems can now draw meaningful statistical inferences pertinent to each individual from massive data sets that include genomic data, imaging results, and biomarker analyses along with traditional clinical variables. Such evidence, made available to clinicians working at the point of care, can direct the most appropriate preventive and therapeutic actions”. The authors of this statement included: Sanders Williams, Dean of Medicine at Duke University; Hunt Willard, Director of the Institute for Genome Sciences and Policy, a distinguished human geneticist and member of the Secretary of Health and Human Services Advisory Committee on Genetics, Health and Safety; and Ralph Snyderman, the Chancellor of the University. They challenged other medical centers to adopt a personalized care model targeting the care of high-risk individuals, and reducing the cost of health care. Larger health care centres and organizations treat patients with uncommon and common chronic diseases. Patients’ response to therapy may be determined by genetic factors just as their likelihood of disease and the severity of disease are also genetically determined. Clinical research now has access to an array of genetic tools to study of complex or common traits, and our challenge as clinicians will be to use these tools to improve the management of patients.
The US Secretary of Health’s report on Pharmacogenomics (see web resources) estimates that by 2020 there will be 134 million Americans with chronic disease. Further a small percentage of the US population represents the majority of health care costs and these costs are directed primarily towards the care of chronic disease. The report expressed concern that effective treatments for chronic disease are under-utilized. Fifty percent of patients on chronic treatment discontinue therapy after 1 year. The future may try the application of genetic tools to improve the care of these patients.
THE IMPORTANCE OF FAMILY HISTORY
The US Surgeon General has undertaken to educate the public on the importance of genetic make-up and the health risks one may encounter in a life time as a result of one’s genetic constitution. A free web-based tool is available for anyone in both English and Spanish, to collect a family history by accessing the web. In preparing such a document, a patient can then discuss potential genetic risks with their health care provider. If a patient has a first degree relative (mother, father, sister, brother) with a common disease, they have a 2-3 fold higher risk of developing the same disease as compared to the general population [4].
In a not uncommon scenario, a patient in his 50’s consults you about his risk of glaucoma, as he knows that his mother has it. You are able to obtain a family history; however, as this is a late-onset disorder, there are no other family members who can be identified by history with glaucoma that allow you to identify a pattern of inheritance (Fig. 1). What information, either from your general knowledge of the genetics of glaucoma, or empirically from the literature, is available to assist? Historically, you may have depended on his racial origin (with genetic implications) to assist you in suggesting a genetic risk. For example, if your patient is of Chinese ancestry, he may be at higher risk for angle-closure vs open-angle glaucoma, or if he is of Afro-Caribbean descent, he may be at increased risk for primary open-angle glaucoma. If he has moderate or high myopia he may have an increased risk of POAG [5]. If he had been exposed to steroids, he might be more at risk for glaucoma. His age will also be a factor to consider. In Canada, 2.7% of individuals over 40 and 11% over 80 have glaucoma [6].
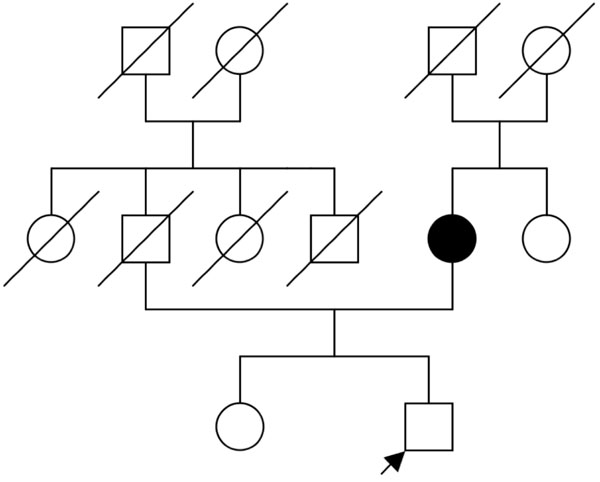 |
Fig. (1). A family history of glaucoma. |
SINGLE GENES AND GLAUCOMA
In some cases a specific gene or locus is associated with glaucoma (Table 1). For example, mutations in single genes have been associated with early-onset primary open angle glaucoma (POAG), congenital glaucoma, and anterior segment dysgenesis. A locus1 suggesting a single gene has been found for pigment dispersion syndrome [7]. In some cases of glaucoma, for example, adult-onset primary open angle glaucoma and normal tension glaucoma, multiple factors combine to create the glaucoma phenotype. For pseudoexfoliative glaucoma, a polymorphism2 in a single gene appears to act as a disease modifier [8].
Single Genes Causing or Modifying the Severity of Glaucoma
| Locus | Condition | Gene |
|---|---|---|
| 1q24-q25;2p21;5q21-q22;10p15-p14 | Early, Adult-onset POAG;Low tension glaucoma | MYOC;CYP1B1;WRD36;OPTN |
| 2p21 | Congenital glaucoma | CYP1B1 |
| 4q25;6q25 | Axenfeld-Rieger syndrome; Iridogoniodysgenesis | PITX2;FOXC1 |
| 9q34 | Nail patella syndrome | LMX1B |
| 11p13 | Aniridia | PAX6 |
| 15q22 | Pseudoexfoliation | LOXL1 |
SNPs and Candidate Genes
| Gene | Drug | Population |
|---|---|---|
| PG F2-αreceptor | latanoprost | Japan [24] |
| β1- adrenergic receptor | betaxolol | USA (48 volunteers) [25] |
| β2-adrenergic receptor | β-blocker | USA (215 glaucoma ) [26] |
| β2-adrenergic receptor | timolol | Austria [27] |
| CYP2D6 | timolol | Finland (19 glaucoma,18 normal) [28] |
Myocilin, previously known as TIGR (for trabecular meshwork inducible), was the first glaucoma gene to be isolated and has been associated with both juvenile open angle glaucoma and adult-onset glaucoma [9]. The WDR36 gene, a member of the WD40 repeat protein family, may be involved in T-cell activation. T cell-mediated responses have been hypothesized to participate in glaucoma-associated optic nerve degeneration. The segregation of a mutation in this gene was found in with a large 5-generation family with POAG by Monemi and colleagues [10]. The association of changes in WDR36 with glaucoma was not replicated in matched cohorts of patients with POAG and normal controls [11].
The OPTN gene encodes a protein called optineurin that interacts with huntingtin and Rab8 [12] and is upregulated in response to sustained elevation of intraocular pressure, TNF-α exposure and dexamethasone, suggesting that it has a protective role in the trabecular meshwork. Mutations in OPTN appear to be clustered in patients with low tension glaucoma (LTG) [13]. One non-synonymous3 mutation, a single base change gaunine (G) to adenine (A) at position 458 of exon 44, replaces the amino acid glutamic acid with a lysine. This particular mutation appears to confer a more severe phenotype of LTG and was not found in the cohort of patients with POAG. Together, mutations in the MYOC, WDR36 and OPTN genes explain only 10% of sporadic cases of POAG [14]; and so, whereas testing for mutations in genes such as MYOC is available on a clinical basis, without a family history of glaucoma, the yield will be low and may not affect the management of the patient.
Congenital glaucoma caused by mutations in the CYP1B1 gene is an autosomal recessively inherited trait. The CYP1B1 gene is involved in the initial phase of metabolism of drugs and the metabolism of endogenous steroids, retinoic acid, arachidonic acid, and melatonin as reviewed by [15]. Whereas many mutations in the CYP1B1 gene have been correlated with glaucoma, the direct links with specific metabolic pathways remain speculative. The gene has three exons. Mutations are commonly found in exons 2 and 3 [15] and clinical testing for mutations is available [16]. Mutations in CYP1B1 may also act as genetic modifiers in cases of POAG due to mutations in the MYOC gene [17]. As reviewed by Vasiliou and colleagues [15], mutations in CYP1B1 occur in a small but important percentage of cases of POAG in geographically separated and likely genetically distinct populations.
The genes PITX2 and FOXC1 encode transcription factors that are both implicated in the development of the anterior segment of the eye. Mutations in these genes result in Axenfeld-Rieger syndrome and iridogoniodysgenesis (also termed iris stromal hypoplasia) [18]. The LMX1B gene encodes a LIM homeodomain containing protein that is strongly expressed in the dorsal mesenchyme and anterior or distal limb structures mirroring the developmental defects seen in the Nail-patella syndrome. The classic transcription factor involved in the development of the eye, PAX6, is associated with aniridia and secondary glaucoma [19].
LOXL1 is the gene encoding the protein lysyl oxidase-like 1. This protein is seen in areas of elastogenesis and interacts with fibulin-5. Single nucleotide polymorphisms (SNPs) in this gene have been associated with pseudoexfoliative glaucoma [8] and further, haplotypes have been defined which predispose to or protect against glaucoma in patients with pseudoexfoliation syndrome [20]. Polymorphisms in LOXL1 appear not to be associated with POAG [21] and further do not appear to confer susceptibility to pigment dispersion syndrome or pigmentary glaucoma [22].
GENES AS DISEASE MODIFIERS AND FACTORS IN DRUG RESPONSIVENESS
Drugs of certain classes are recognized as being more effective in certain racial groups and by implication in a genetic group. The basis for these clinical responses is now being investigated with genetic tools using single nucleotide polymorphisms (SNPs) for candidate genes [23]. From clinical experience, some patients respond very well to latanoprost with intraocular pressure lowering whereas others do not. The poor-responders may in turn respond well to travaprost or bimatoprost. A hint as to the genetic variability that underlies drug response was provided by a study by Sakurai and colleagues [24] in which, the IOP lowering response of latanoprost was correlated with a SNP in the prostaglandin (PG) F2-α receptor gene in 100 Japanese normal volunteers. Other similar studies have correlated SNPs in genes encoding drug receptors and genes involved in drug metabolism with clinical efficacy of glaucoma medications (Table 2). At present these reports have not yet resulted in a commercially available test which can be applied to patient care.
GENETICS AND INSIGHTS INTO DISEASE MECHANISMS
The identification of genes and the creation of animal models are helping to elucidate the physiological pathways that ultimately cause glaucoma [29]. The tools of genomics, molecular biology, developmental biology, bioinformatics and computational biology are all contributing to a greater understanding of these pathways. This knowledge should ultimately lead to a better understanding of the normal physiology of the trabecular meshwork, optic nerve, ganglion cells, and other associated tissues. An improved understanding of the state of the eye in disease and health will facilitate the rational development of drugs tailored to specific subtypes of glaucoma.
PRIVACY AND COUNSELLING
Legislation to protect against genetic discrimination is important. In response the perceived lack of protection of patients’ rights and the growing ability of genetics to type individuals as affected or at risk, and after many years of lobbying, the Genetic Information Non-discrimination Act (GINA) was signed into law in the USA. The Act offers protection against discrimination in applications for health insurance and employment based on genetic information [30].
Clinicians need to safeguard genetic data, and also ensure appropriate genetic counselling is available. The role of a genetic counsellor is to be an informer (not an advisor) providing facts and options, so that an informed decision can be made by the patient and his/her caregiver(s).
SUMMARY
We are entering an era of genomic medicine, where genetic tools are available to improve care of patients and make best use of health care expenditures. Enlightened patients, aware of their family history, will ideally seek care earlier to avoid the complications of chronic diseases such as glaucoma. Physicians who understand genetic risk can develop a personalized prescription of care targeting at risk individuals and tailoring their care towards efficacy of interventions while minimizing adverse reactions to treatments. As practionners we look forward to the availability of genetic tests for our patients, and will need to be mindful about how this information is protected and shared. At the same time we realize that we as practitioners will need to be educated in how these tests can be applied and how we can translate their results to improve the care of our patients.
WEB-BASED RESOURCES
Realizing the potential of pharmacogenomics: Opportuities and challenges. Report of the Secretary’s Advisory Committee on Genetics, Health and Society. May, 2008. http://oba.od.nih. gov/oba/SACGHS/reports/SACGHS_PGx_report.pdf
US Surgeon General’s Family History Initiative. http://www.hhs.gov/familyhistory/
NOTES
1 A locus is the location of a gene on a chromosome.
2 A polymorphism is a heritable change in the DNA base sequence, generally common.
3 A non-synonymous change in the genetic code results in the replacement of an amino acid by another amino acid. A synonymous change occurs when the base change but does not result in an amino acid change, as there is some redundancy in the genetic code.
4 The coding sequences of a gene are generally referred to as exons. DNA sequences between the exons are termed intervening sequences or introns and, in general, these do not normally form part of the messenger RNA that forms the protein product of a gene.
ACKNOWLEDGEMENTS
I sincerely appreciate the advice of two experts in glaucoma: Dr. Ordan Lehmann and Dr. Karim Damji who reviewed the manuscript and provided important suggestions.






