All published articles of this journal are available on ScienceDirect.
Molecular Variants in Genes related to the Response to Ocular Hypotensive Drugs in an Afro-Colombian Population
Abstract
Aims:
This study aimed to conduct an exploratory analysis of the pharmacogenomic variants involved in ocular hypotensive drugs to understand the individual differential response in an Afro-descendant population.
Background:
Glaucoma is the leading cause of irreversible blindness worldwide. The pharmacologic treatment available consists of lowering intraocular pressure by administering topical drugs. In Asian and Caucasian people, pharmacogenomic variants associated with the efficacy of these treatments have been identified. However, in Afro-descendant populations, there is a profound gap in this knowledge.
Objective:
This study identified the pharmacogenomic variants related to ocular hypotensive efficacy treatment in Afro-descendant individuals from the Archipelago of San Andres and Providence, Colombia.
Methods:
An analysis of whole-exome sequencings (WES), functional annotation, and clinical significance was performed for pharmacogenomic variants reported in PharmGKB databases; in turn, an in silico available prediction analysis was carried out for the novel variants.
Results:
We identified six out of 18 non-synonymous variants with a clinical annotation in PharmGKB. Five were classified as level three evidence for the hypotensive drugs; rs1801252 and rs1801253 in the ADRB1 gene and rs1042714 in the ADRB2 gene. These pharmacogenomic variants have been involved in a lack of efficacy of topical beta-blockers and higher systolic and diastolic pressure under treatment with ophthalmic timolol drug. The rs1045642 in the ABCB1 gene was associated with greater efficacy of treatments with latanoprost drug. Also, we found the haplotypes *17 for CYP2D6 and *10 for CYP2C19; both related to reducing the enzyme activity to timolol drug metabolization. In addition, we observed 50 novel potentially actionable variants; 36 synonymous, two insertion variants that caused frameshift mutations, and 12 non-synonymous, where five were predicted to be pathogenic based on several pathogenicity predictions.
Conclusion:
Our results suggested that the pharmacogenomic variants were found to decrease the ocular hypotensive efficacy treatment in a Colombian Afro-descendant population and revealed a significant proportion of novel variants with a potential to influence drug response.
1. INTRODUCTION
Glaucoma is the leading cause of irreversible blindness worldwide. It is a group of optic neuropathies characterized by changes in the optic nerve head and typical visual field defects secondary to a progressive loss of retinal ganglion cells. The etiology of this disease is not entirely known. Glaucoma is usually considered a multifactorial disease involving genetic, vascular, inflammatory, and oxidative factors in its pathogenesis. The most critical risk factor is elevated intraocular pressure (IOP), although this is not enough or required for disease development. Other risk factors include advanced age, Afro-descendant or Hispanic ethnicity, positive family history, and high myopia. The available therapies are based on reducing intraocular pressure through medications or surgery. Most patients are treated with topical ophthalmic drugs, including first-line prostaglandin-analogs such as latanoprost, travoprost, and bimatoprost, followed by beta-adrenergic blocking agents (e.g., timolol is the most representative) [1]. Intraocular pressure response rates to these drugs are known to vary according to the population analyzed. For example, Camras and Hedman [2] conducted a comparative study between latanoprost and timolol in the North American population and found null-response rates of 20% and 31%, respectively; furthermore, these rates decreased by 4.7% and 1.6% after four months. Other studies that evaluated only latanoprost across different periods reported null-response rates of 4.1% in the Italian population [3], 11% in the Spanish population [4], and 31.8% in the Japanese population [5].
Glaucoma disproportionally affects Afro-descendant populations at a frequency three to five times greater than European populations, and its onset has been reported at a younger age, with a more severe course of the disease. However, there is a gap in knowledge given the lack of studies on Afro-descendant populations that contribute to understanding the susceptibility and adverse outcomes in this ethnic group. Some reports point to a differential response to hypotensive drugs. There was an observed inefficacy of topical beta-blockers and rebound hypertension in healthy volunteers in Nigeria. Other reports suggest a better therapeutic response to travoprost than to latanoprost or timolol in Afro-descendant patients [6-11].
Identifying molecular variants associated with drug responses is essential in precision medicine since this helps maximize efficacy and minimize toxicity for each patient [12-14].
This research aimed to conduct a cross-sectional study to identify molecular variants in genes associated with the response to hypotensive drugs for primary open-angle glaucoma (POAG) in Afro-descendant individuals. We hypothesized that pharmacogenetic variants are associated with differential responses to ocular hypotensive drugs in Afro-Colombian individuals.
2. MATERIALS AND METHODS
A cross-sectional study was conducted to analyze molecular variants in genes associated with the response to and metabolism of the ocular hypotensive drugs latanoprost and timolol. For this, bioinformatics analysis was performed on ten whole-exome sequences of Afro-descendant Raizal individuals from the Archipelago of San Andres and Providence, Colombia, in August 2019.
The study sample size was determined based on the participants' signed informed consent for the project “Analysis of exomes of patients with glaucoma in the Raizal population of the archipelago of San Andres and Providence” and the ethical approval act #106-019 granted by the Human Ethics Institutional Review Committee (CIREH) of the Faculty of Health of Universidad del Valle. The study was conducted according to the Helsinki Declaration, as revised in 2013.
Functional annotation and clinical significance were performed for the variants reported in databases; in turn, an in silico available prediction analysis was carried out for the non-evaluated variants. The frequencies of these variants were compared with those present in other populations worldwide. Molecular variant annotation was performed based on alternative allele frequency (AAF), type of variation, clinical annotation, and functional prediction.
2.1. DNA Extraction and Sequencing
Peripheral blood samples were obtained through finger-pricking. According to the manufacturer's instructions, DNA extraction was done using the QIAmp® DNA Blood Maxi kit (QIAGEN®). Exome sequencing was done on DNA samples that met the following quality criteria: concentration >10 ng/µl and A260/280 ratio of 1.7 by spectrophotometry on a Nanodrop® ND-2000c (Thermo Scientific®). Exome capture and enrichment were done using the SureSelect V6 library preparation kit, and sequencing was performed on Illumina NovaSeq 600 equipment at 100X depth to address potential sources of bias.
2.2. Read Mapping and Molecular Variant Calling
Read quality control was evaluated using FastQC software [15]. Reads with more than 20% of bases with a quality score below 30 were discarded. The short reads were mapped to the GRCh38 reference genome using BWA-mem. SAM to BAM format conversion was done using GATK 4.1.6 [16], followed by SNP variant calling using HaplotypeCaller. Next, BED tools were used to filter variants in candidate genes [17], which were selected based on a literature review and database search focused on potentially associated variants according to the Pharmacogenomics Knowledge Base (PharmGKB) [18].
2.3. Annotation and Functional Prediction
Molecular variant annotation was performed based on alternative allele frequency (AAF), type of variation, clinical annotation, and functional prediction, among other information obtained from WANNOVAR [19]. The haplotypes and alleles for the CYP2D6 gene were manually assessed by visualizing the VCF file on GenomeBrowse and comparing the data against PharmVar [20]. An allele frequency was calculated by dividing the number of times the allele of interest was observed in a population by the total number of copies of all the alleles at that genetic locus in the population. The CADD server was used to predict potentially deleterious molecular variants (value >20). Furthermore, the SIFT, PolyPhen, and Mutation Assessor server (https://useast.ensembl.org/info/genome/variation/prediction/protein_function.html) were also used.
2.4. Copy Number Variation
Copy number variation (CNV) for the CYP2D6 gene was determined using the BAM file as input [21].
3. RESULTS
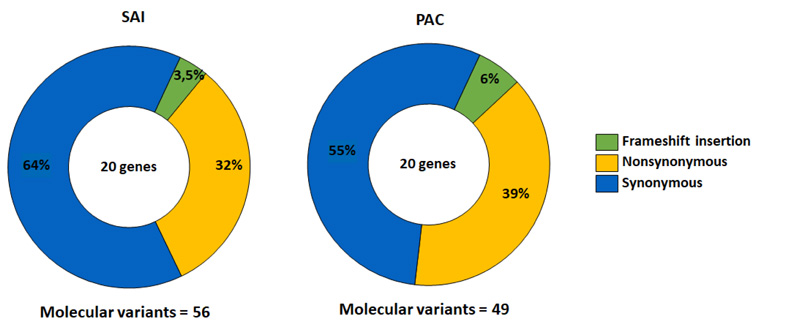
From the 10 participants, 56 molecular variants were identified in 20 analyzed genes. 36 variants were synonymous, followed by 18 non-synonymous, and a small number of insertion variants caused frameshift mutations [2] (Fig. 1).
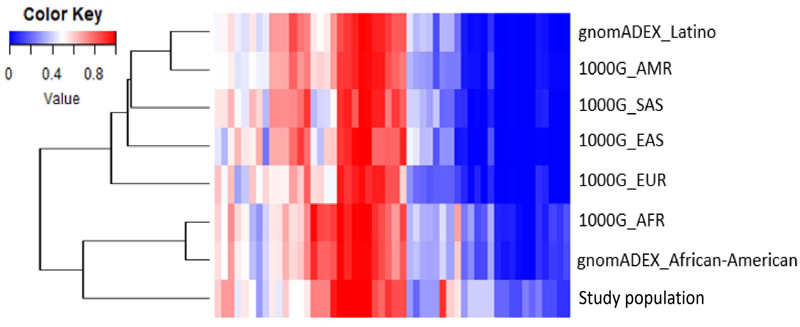
The AAF of the molecular variants was compared with the data of seven populations from the 1000 Genomes Project and gnomAD. The results are shown by a heatmap and dendrogram reconstructed using Euclidean distances as similarity metrics (Fig. 2). The clustering pattern indicates that the study samples are similar to the populations of African descent, showing a clear separation from the others.
The non-synonymous molecular variants with no reported clinical associations were scored according to five in silico functional prediction servers (Table 1). The CADD server identified five potentially deleterious molecular variants (value >20). Furthermore, the SIFT server predicted three deleterious molecular variants, PROVEAN and PolyPhen servers identified two, and MutationAssessor predicted one variant.
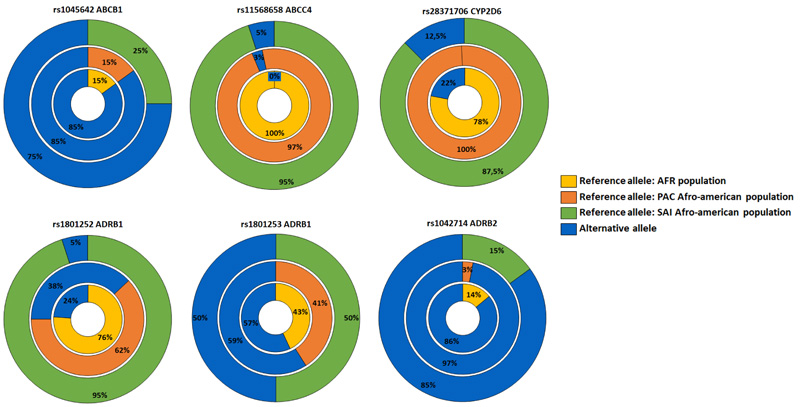
A total of six variants with reported clinical association were found in five of the genes evaluated, including three variants associated with increased drug efficacy, two with reduced drug efficacy, and one with toxicity (Table 2). Compared to the reference populations, these variants showed lower frequencies of the alternative alleles associated with differential therapeutic responses in all variants except for the ABCC4 gene (Fig. 3).
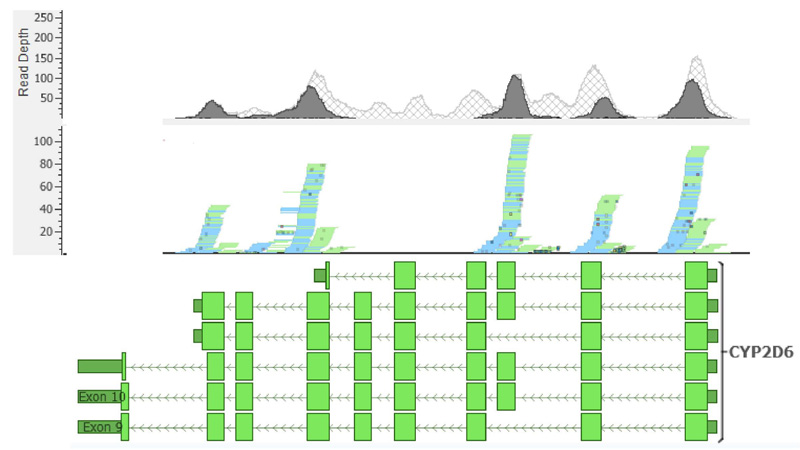
The assignment of metabolic phenotypes for the CYP2D6 gene was not possible due to ambiguous read mapping in certain gene regions containing the alleles that define the haplotypes of metabolic phenotypes. Likewise, copy number variant calling was also affected since the algorithm used in CONIFER depends on calculated depth values for different exons; therefore, ambiguous read mapping can lead to incorrect conclusions on the presence of multiple copies in the samples analyzed. The read assignment pattern was constant across samples (Fig. 4). Exons 4 and 8 of the CYP2D6 gene showed shallow read depth, and for exons 5 and 6, no reads could be assigned.
| Chromosome | Position | Ref | Alt | Gene | dbSNP | SAI Alt Frequency | PAC Alt Frequency | SIFT Score/Pred | Mutation Assessor Score/Pred | PROVEAN Score/Pred | Polyphem Score/Pred | CADD Pred |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 161193676 | G | A | ADAMTS4 | rs147127522 | 0.15 | 0 | 0.0/D | 3955/H | -6.54/D | 1.0/D | 34 |
| 10 | 94781858 | C | T | CYP2C19 | rs6413438 | 0.25 | 0 | 0.007/D | 3375/M | -8.8/D | 1.0/D | 26.1 |
| 19 | 41352971 | C | G | TGFB1 | rs1800471 | 0.15 | 0.15 | 0.252/T | 1.2/L | -0.5/N | 0.012/B | 26.3 |
| Gen | Variant | Type of Interaction | Drug | Level of Evidence | SAI Alt Frequency | PAC Alt Frequency |
|---|---|---|---|---|---|---|
| ABCB1 | rs1045642 | Efficacy | Latanoprost | 3a | 0.75 | 0.85 |
| ABCC4 | rs11568658 | Inefficacy | Latanoprost | 3 | 0.05 | 0.03 |
| ADRB1 | rs1801252 | Inefficacy Toxicity | Timolol | 3 | 0.05 | 0.38 |
| ADRB1 | rs1801253 | Efficacy | Beta-blocking agents | 3 | 0.5 | 0.59 |
| ADRB2 | rs1042714 | Efficacy | Timolol | 3b | 0.85 | 0.97 |
| CYP2D6 | rs28371706 | Substrate specific | - | 1A-4 | 0.13 | 0.00 |
SAI = San Andres Island population, PAC = Pacific colombian population, ALT = Alternative allele
4. DISCUSSION
The Colombian population comprises Amerindian, European, and African descent [22]. Some genetic studies on the population, such as the one conducted by Lamprea [23], indicate that the Raizal populations of the San Andres and Providence islands are more like the Caribbean Anglo-speaking population. This can be expected since these populations share a history of slavery and colonization by Great Britain in the different islands of the Caribbean [24]. Our results agree with this fact and can be observed by the clusters generated between African and Raizal study populations. The absence of Latin and Anglo-American people in the cluster analysis was due to the limited molecular data on the Colombian people in the 1000 Genomes project. Currently, the only data available corresponds to a population of primarily European origin from Antioquia in the Colombian Andean region [25, 26].
Based on the complete exome characterization, we evaluated non-synonymous variants since these can affect protein function by active site modifications or structural destabilization [27]. These molecular variants accounted for 32% of the total variants identified (Fig. 1).
In silico functional prediction analysis using different bioinformatics servers was done to establish the possible impact of the molecular variants on the encoded proteins [27, 28]. Five out of six clinical annotations in PharmGKB were classified as level 3 evidence for the studied hypotensive drugs. These associations are not robust enough for clinical applications since they were based on a single unreplicated study on variant-drug combinations [29].
Regarding the possible effects of the molecular variants identified in genes associated with differential drug response, we found that variant rs1801252 (A>G) in the ADRB1 gene is involved in a lack of efficacy of topical beta-blockers and higher systolic and diastolic pressure under treatment with ophthalmic timolol [30]. The adrenergic receptors ADRB1 and ADRB2 belong to a large superfamily of G-protein-coupled receptors and are the main targets of beta-adrenergic antagonists [31]. Moreover, for variants rs1801253 in ADRB1 (G>C) and rs1042714 (G>C) in ADRB2, homozygous carriers (CC) have been associated with a higher probability of intraocular pressure reduction with beta-blockers [31, 32]. In the Raizal population of San Andres and Providence, the homozygote frequencies were 0.3 and 0.7, respectively. All molecular variants observed for these genes were found at a lower frequency than in Africans; therefore, these should be considered when evaluating the differential response to timolol in the study population.
ATP-dependent transporters (ABC) are associated with various endogenous and exogenous molecules of different nature. ABC transporters have been suggested as a potential early biomarker in glaucoma due to their involvement in chronic vascular dysregulation, a common phenomenon in these patients [33]. These transporters are crucial in drug efflux; they mediate multi-resistance processes and differential responses to many drugs. In the study group, two molecular variants were detected; rs1045642 (C<T) in ABCB1 and rs11568658 in ABCC4 (C>T). The variant in the ABCB1 gene is associated with greater efficacy of treatments with latanoprost, while its genotype TT confers greater susceptibility to POAG [34].
Our results showed a lower minor allele frequency (0.75) than in the African population (0.85). The homozygous genotype TT was present in 50% of the study individuals. Moreover, genotype CT of the variant in the ABCC4 gene has been associated with a reduced latanoprost effect compared to genotype CC [35]. In this study, a single heterozygous individual carried the variant, which agrees with its low global frequency worldwide. It has not been reported in the African population, according to data from the 1000 Genomes Project. Despite the proven importance of ABC genes in pharmacokinetics, some authors propose that the impact of molecular variants on the bioavailability of drugs is lower than other factors that regulate the expression and function of the transporter [36]. Additionally, most studies on currently annotated molecular variants do not strongly support these associations; therefore, new studies are needed to propose novel variants and discard or keep the existing ones.
P450 cytochrome (CYP) enzymes play a critical role in the metabolism of a broad spectrum of drugs, including proton pump inhibitors, antiepileptic agents, chemotherapeutic agents, and beta-adrenergic antagonists. Timolol is locally well-tolerated; however, up to 80% of the eye drop topically applied can reach systemic circulation [37] and potentially cause cardiovascular and respiratory side effects. Therefore, a normal function of the enzymes involved in their metabolism is highly relevant, especially those associated with drug kinetics and plasma concentrations. CYP2D6 and CYP2C19 are the main metabolizers of timolol [38]. These enzymes are characteristically highly polymorphic, causing changes in their expression levels and enzyme selectivity and activity. The two molecular variants found in these genes, *17 for CYP2D6 and *10 for CYP2C19, belong to the haplotypes known to reduce the enzyme activity. rs2871706 in the CYP2D6 gene displays substrate-specific effects and can reduce protein function depending on the molecule it binds to [39]. Moreover, the variant predicted by CADD for CYP2C19 (rs6413438) has been associated with a deleterious effect on the expression of the enzyme [40]. Both variants have been related to populations of African descent [41-44]; therefore, clinical studies must be conducted to understand the role of these variants in timolol metabolism and their implications in differential drug responses in these populations.
The characterization of the CYP2D6 gene in this study presented several challenges due to the inherent limitations and proposed methodology. First, WES excludes non-coding regions, which are often essential to define haplotypes and metabolic phenotypes; therefore, the results cannot be considered conclusive. Furthermore, the complex structure of the CYP2D6 gene region is challenging for short-read sequencing technologies, given the existence of pseudogenes, repetitive elements, high polymorphism density, etc [45]. These elements cause ambiguous reading assignments since reads can map to other homologous gene regions or no reading assignment because of high alignment penalties and high polymorphism rates. Overall, these issues can lead to biased and inaccurate variant calling [46] that hinders the assessment of structural variants, such as CNV, through reading depth-based algorithms. Consequently, the inconsistencies in reading coverage and depth observed in our data are not unexpected (Fig. 4) and have been previously reported in other WES-based studies [47].
Most molecular variants with functional impact were found in genes that are associated with the downstream cascade of latanoprost action despite not directly participating in drug kinetics or dynamics. These genes are related to extracellular matrix remodeling and degradation; the latter is essential to the mechanism of action of latanoprost to increase the aqueous humor outflow through an unconventional (uveoscleral) route; degradation of the extracellular matrix between ciliary muscle fibers [48]. The variant in the MMP17 gene obtained a high prediction value by CADD. This matrix metalloproteinase activates aggrecanase 1 (ADAMTS4), which plays an essential role in the degradation of glycosaminoglycans of the extracellular matrix [48, 49]. A variant in ADAMTS4 was also predicted to affect its protein function. In addition, a variant with the prediction of functional impact was identified in the TGF-β1 gene, which has been associated with an imbalance between matrix-degrading enzymes (metalloproteinases) and their inhibitors (TIMP), promoting an extracellular stiffness matrix at the outflow pathways [50]. Lastly, a variant with a possible functional impact on EGFR could be associated with side effects, such as trichomegaly, after the administration of latanoprost because the drug-receptor interaction of latanoprost with ADRB can cause transactivation of EGFR receptors [51]. Therefore, a change in protein function can lead to secondary dermatological effects [52].
Despite representing the most remarkable genomic diversity globally, African communities are strongly underrepresented in genomic databases, and clinical research on these populations is limited [53]. This leads to significant limitations and challenges in developing and interpreting studies such as this one. As previously mentioned, for CYP2D6, read mapping to a reference genome can introduce a bias that underestimates the actual variation in the samples. This is because the alignment algorithm systematically penalizes high genetic diversity between the sample and reference genomes, termed allelic or reference bias [54]. In the variant call, this type of bias can lead to calling variants only because the reference genome displays a rare allele or, conversely, not calling rare alleles because the reference shares them. This bias can lead to emphasizing the reference genome's properties instead of the widespread properties across the population [55]. The alternatives proposed to overcome these biases include ethnicity normalized reference genomes [56] and consensus genomes that seek to reflect the most common alleles and molecular variants by replacing positions where the reference contains rare alleles. Another alternative involves graphic genomes [57], representing all possible polymorphisms in a single reference genome. On this basis, studies such as ours that describe and characterize variation contribute substantially to this type of alternative and facilitate genomic studies in these populations.
CONCLUSION
In conclusion, this study identified variations in the main pharmacogenomics associated with anti-glaucomatous therapy in an Afro-Colombian Raizal population. Our results suggested that the pharmacogenomic variants were found to decrease the ocular hypotensive efficacy treatment in a Colombian afro-descendant population and revealed a significant proportion of new variants with a potential to influence drug response. Also, we described the characteristics of the sample regarding the frequency of its molecular variants compared to others based on information in public databases. This comparison allowed determining ethnicity-specific associations of the biomarkers analyzed in the study population. Furthermore, we highlighted several molecular variants that have not been evaluated for hypotensive drugs but could be contributing to differential drug responses in these communities. These findings provide opportunities for future association studies. This study contributes to the Raizal community's genomic data and knowledge of glaucoma pharmacogenetics.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The ethical approval act #106-019 was granted by the Human Ethics Institutional Review Committee (CIREH) of the Faculty of Health of Universidad del Valle.
HUMAN AND ANIMAL RIGHTS
No animals were used that are the basis of this study. All the human experiment were performed in accordance with the Helsinki Declaration, as revised in 2013.
CONSENT FOR PUBLICATION
Informed consent was obtained from the participants.
STANDARDS OF REPORTING
STROBE guidelines were followed.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
The vice-rector financially supported the research (CI 71215), and AC was supported by a US-National Institutes of Health grant R01 NS110122
CONFLICT OF INTEREST
The authors declared no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors especially thank Nathalie Abraham for helping with the presentation of the study to the Raizal population in their native language and Andrea Gonzalez for English editing.


